Introduction
Hello AI Enthusiasts, In this post, we will learn how to apply 3-D CNN with Keras. For the dataset, we will use the MNIST 3-D available at Kaggle. Link.
Before that let's try to understand the working of 3-D CNN,
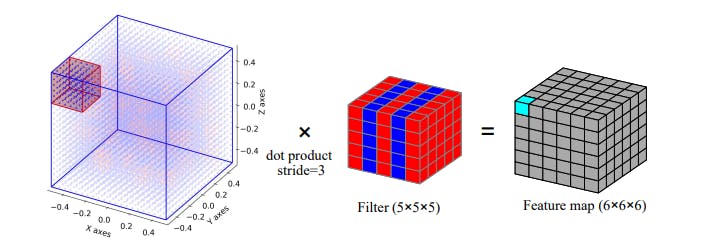 Image Credit - Arxiv paper - 3D-CNN for heterogeneous material homogenization
Image Credit - Arxiv paper - 3D-CNN for heterogeneous material homogenization
In 3D-CNN, we simply switch from 2-D to 3-D. So, now our kernel is 3D, pooling is 3-D and the resulting FeatureMaps are also 3-D.
Another change is the convolution steps, the filter convolutes the input in 3-D space instead of just a 2-D plane. The first step of the 3-D convolution is depicted in the above image. You can extrapolate it in a 3-D space.
In 2-D also, the FeatureMaps along with the channels used to be a 3-D but in 3-D CNN individual FeatureMaps are 3-D if we consider the channels (i.e. result of each Kernel) together, it will be a 4-D.
Need of 3-D CNN
The immediate question that may arise is what's the purpose of 3-D CNN and how it will be better than a 2-D CNN.
A simple answer is, just like the way a 2-D facilitates spatial invariance in a 2-D plane, a 3-D CNN will facilitate spatial invariance in a 3-D space.
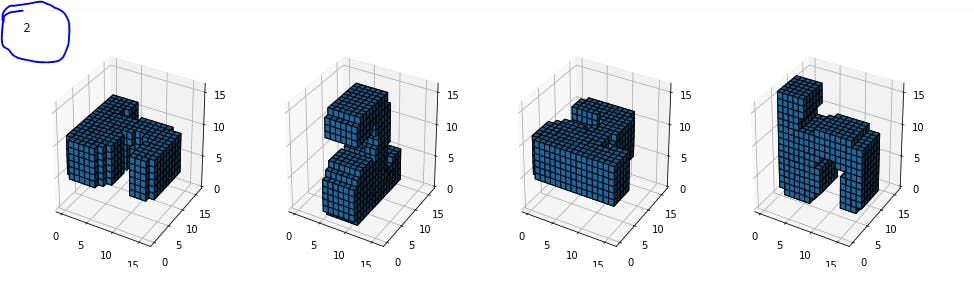
Let's check these MNIST digits,
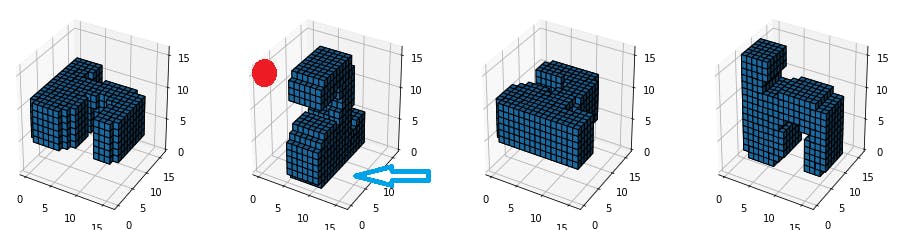 If we observe the highlighted digits(2nd), it resembles a "2" even with a 2-D slice from the front but this 2-D slice will no work if the image is rotated in a 3-D space(see other images). What we need here is to capture the 3-D features.
If we observe the highlighted digits(2nd), it resembles a "2" even with a 2-D slice from the front but this 2-D slice will no work if the image is rotated in a 3-D space(see other images). What we need here is to capture the 3-D features.
This was just one use-case when we add another dimension in 2-D image and made it a 3-D, but there is another use-case of 3-D CNN i.e. adding a temporal dimension in a 2-D frame(a video).
Check this image,
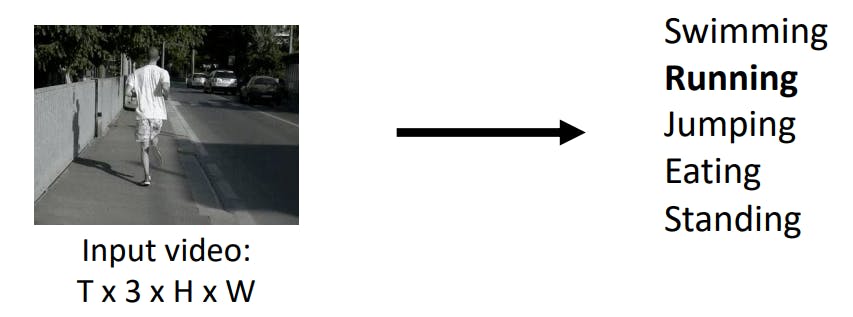 Image Credit - Deep Learning for Computer Vision, University of Michigan
Image Credit - Deep Learning for Computer Vision, University of Michigan
In the above video, we need to convolute the image across the time frame to extract the Feature maps which can distinguish "Running" from "Jumping" by appropriately registering the movement of legs and hands across frames.
Let's code it
We will now jump to the code to implement a 3-D CNN using Keras on MNIST digit dataset.
path = "/content/......./full_dataset_vectors.h5.zip"
with ZipFile(path, 'r') as zip:
zip.extractall("/content/")
with h5py.File("/content/full_dataset_vectors.h5", "r") as hf:
x_train = hf["X_train"][:]
y_train = hf["y_train"][:]
x_test = hf["X_test"][:]
y_test = hf["y_test"][:]
Code-explanation
We have downloaded the dataset from the Kaggle link. Then simply unextracted the zipfile and open the h5 files into train/test. This code is available on the website
num=np.random.randint(0,1000,1)[0] #1
vox = x_train.reshape(-1,16,16,16)[num] #2
vox_1 = np.ceil(vox).swapaxes(0,2) #3
vox_2 = np.ceil(vox).swapaxes(0,1)
vox_3 = np.ceil(vox).swapaxes(1,2)
fig = plt.figure(figsize=(16,4))
#---- First subplot
ax = fig.add_subplot(1, 4, 1, projection='3d') #4
#---- 2nd subplot
ax1 = fig.add_subplot(1, 4, 2, projection='3d')
#---- 3rd subplot
ax2 = fig.add_subplot(1, 4, 3, projection='3d')

#---- 4th subplot
ax3 = fig.add_subplot(1, 4, 4, projection='3d')
ax.voxels(vox, edgecolor='k')
ax1.voxels(vox_1, edgecolor='k')
ax2.voxels(vox_2, edgecolor='k')
ax3.voxels(vox_3, edgecolor='k')
print(y_train[num]) #5
Code-explanation
#1 - Generating a ranomf number to get one random instance from x_train
#2 - x_train is flattened i.e. shape=(10000, 4096). So we have reshaped it into (10000,16,16,16)
#3 -Swaped the each axis once to view the digit in different orientation
#4 -Plotted the different orientation on different axes of the figure
#5 -Printed the digit's label
Result
x_train = x_train.reshape(-1,16,16,16,1) #1
y_train = pd.get_dummies(y_train) #2
x_test = x_test.reshape(-1,16,16,16,1)
y_test = pd.get_dummies(y_test)
from tensorflow.keras.models import Sequential
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.layers import Dense, Dropout, Flatten, Conv3D, BatchNormalization, MaxPooling3D
Code-explanation
#1 - Reshaped the images to keras format i.e. channe_last(channels=1)
#2 - One Hot Encoded the labels
filters = 64
dropout = 0.5
model= Sequential([
Conv3D(filters, 3, padding='same', activation='relu', input_shape = (16, 16, 16, 1)),
MaxPooling3D(pool_size=2, padding="same"),
BatchNormalization(),
Dropout(dropout),
Conv3D(filters, 3, activation='relu', padding='same'),
#MaxPooling3D(pool_size=2, padding="same"),
BatchNormalization(),
Dropout(dropout),
Conv3D(filters, 3, activation='relu', padding='same'),
#MaxPooling3D(pool_size=2, padding="same"),
BatchNormalization(),
Dropout(dropout),
Conv3D(filters, 3, activation='relu', padding='same'),
BatchNormalization(),
Dropout(dropout),
Conv3D(filters, 3, activation='relu', padding='same'),
BatchNormalization(),
Dropout(dropout),
Flatten(),
Dense(250, activation='relu'),
Dropout(dropout),
Dense(100, activation='relu'),
Dropout(dropout),
Dense(10, activation='softmax')
])
#model.summary()
adam = Adam(learning_rate=0.001)
model.compile(loss='categorical_crossentropy', optimizer=adam, metrics=['accuracy'])
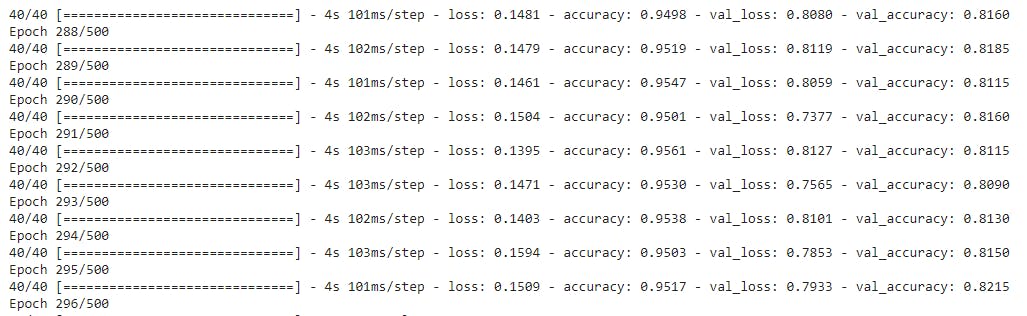
model.fit(x_train, y_train, batch_size=250, epochs=500, validation_data=(x_test, y_test))
Code-explanation
All the code above is self-explanatory, the only thing that is changed is the use of 3-D CNN and 3-D pooling.
Kernels size, Dropout etc. are hyper-parameters and have been found by tuning approaches. Just like we do in a regular 2-D CNN.
Result
Summary
This was all from us for this post. Please try the code in your own setup.
You may try,
- Making a binary classifier by picking only 2 digits
- Try this on any other 3-D dataset
- Go through the slides of University of Michigan